Dr. Stephanie A. Eichorst

One gram of soil contains over a million microorganisms, yet much of their function remains unknown. Stephanie started studying soil microbes during her PhD at Michigan State University, and continued as a postdoc at Los Alamos National Lab, and scientist at the Joint BioEnergy Institute (USA). Since joining the University of Vienna in 2012, she has been investigating soil microorganisms (Acidobacteriota) via cultivation, ‘omics methods and metabolic process measurements. She is currently exploring atmospheric gas oxidation as a means of survival during periods of energy limitation, as well as factors governing the use of different terminal oxidases in soil microbes, which may underpin their physiological flexibility.
Links

Main Research Interests
Physiology of soil microorganisms
Exploring microbial physiology has been a central part of my research for many years, with particular emphasis on members of the Acidobacteriota. Members of the phylum Acidobacteriota are ubiquitous across soils worldwide and comprise a monophyletic bacterial phylum of astonishing diversity.Their common occurrence and high abundance based on ribosomal gene sequences suggest that they are likely a major component of the soil microbial community and play ecologically significant roles in soil processes. Yet, their functions remain largely unknown due to the limited number of cultivated representatives and a paucity of information on their genetic potential (genomic and metagenomic information).
Overall, my goal is to elucidate the ecophysiology and therefore the success and ubiquity of members of the phylum Acidobacteriota in soils. To achieve this goal, we use a combination of genomics, transcriptomics, growth-based experiments, enzyme kinetics and molecular analyses to identify key features in acidobacterial strains. In addition, we continuously thrive to isolate new members of the Acidobacteriota. Some of our investigations are highlighted below.
Shallow breathing

- Several acidobacterial strains harbor both low- and high-affinity terminal oxidase genes.
- Contrary to established dogmas, low-affinity oxidases are expressed under nanomolar oxygen concentrations.
- The presence of high-affinity terminal oxidases in the genome does not per se denote a strain being "microaerobic".
- This respiratory flexibility of the prevalent Acidobacteriota could aid in their survival in the heterogenous soil environment.
- Reference: Trojan et al., 2021. mSystems 6(4):e0025021.
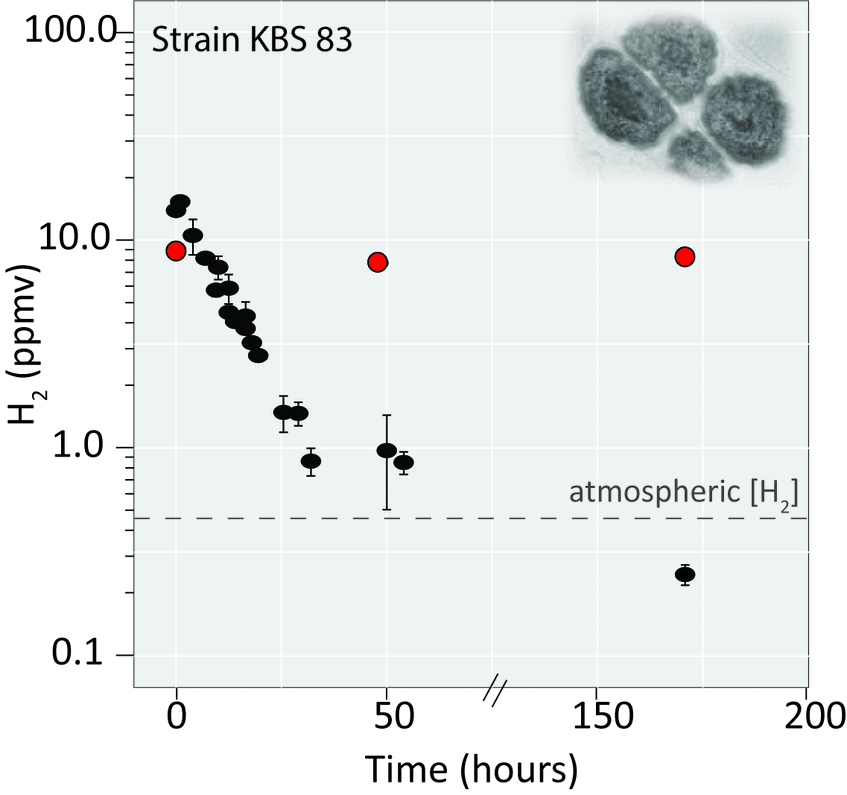
H2-scavenging ability
- We revealed another feature that can explain their success in soil - the existence of group 1h [NiFe]-hydrogenases in several soil acidobacterial genomes.
- We demonstrated the use of this enzyme to scavege molecular hydrogen during carbon starvation in acidobacterial strains.
- Furthermore, we surveyed geographically and edaphically different soils and revealed that Acidobacteriota are prevalent members of the hydrogen-scavening community.
Some of our acidobacterial strains and research have been highlighted here:
- Wie Acidobakterien unter widrigen Bedingungen überleben (sorry, only in German)
- Bacterial survival kit to endure soil.
- Behind the paper
- Bodenmikroorganismen als Überlebenskünstler: Wie schaffen die das? (sorry, only in German)
- Bodenmikroorganismen sind wahre Überlebenskünstler (sorry, only in German)
- Small Things Considered - A Whiff of Taxonomy - The Acidobacteria
Survival mechanisms and dormancy in terrestrial systems
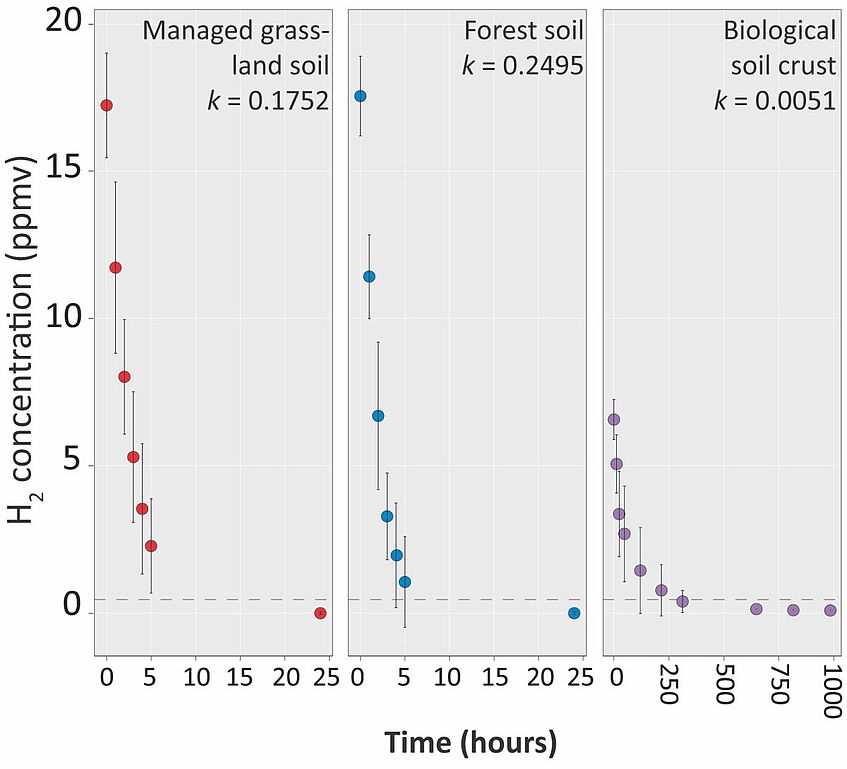
Survival mechanisms and dormancy in terrestrial systems
Terrestrial ecosystems represent habitats with unpredictable conditions for soil microorganisms, often confronting them with suboptimal conditions for growth. As such, up to 80% of microorganisms in soils are assumed to be in a state of non-growth, oftentimes referred to as ‘dormant’ – a state with low metabolic activity enabling long-term persistence. In many soil systems, microorganisms alter between ‘dormant’ and active states, presumably as a means of survival. It is believed that the state of ‘dormancy’ helps to maintain the extensive biodiversity in soils, with estimates ranging from 1,000 to 1,000,000 species per gram of soil.
Atmospheric gas oxidation, such as molecular hydrogen (H2), has been proposed as a means to generate energy during periods of carbon limitation. It is believed to be a widespread survival strategy that contributes to bacterial persistence. It is accomplished due to the activity of high-affinity hydrogenases. We are focusing on two groups of oxygen tolerant high-affinity hydrogenases, groups Ih and 1l [NiFe]-hydrogenases, that are assumed to be major contributors to H2 uptake in soils. Using a combination of genomics, transcriptomics, growth-based experiments, enzyme kinetics and molecular analyses, we are exploring this physiology in model organisms and in temperate and desert soils.
Cultivating the uncultivables
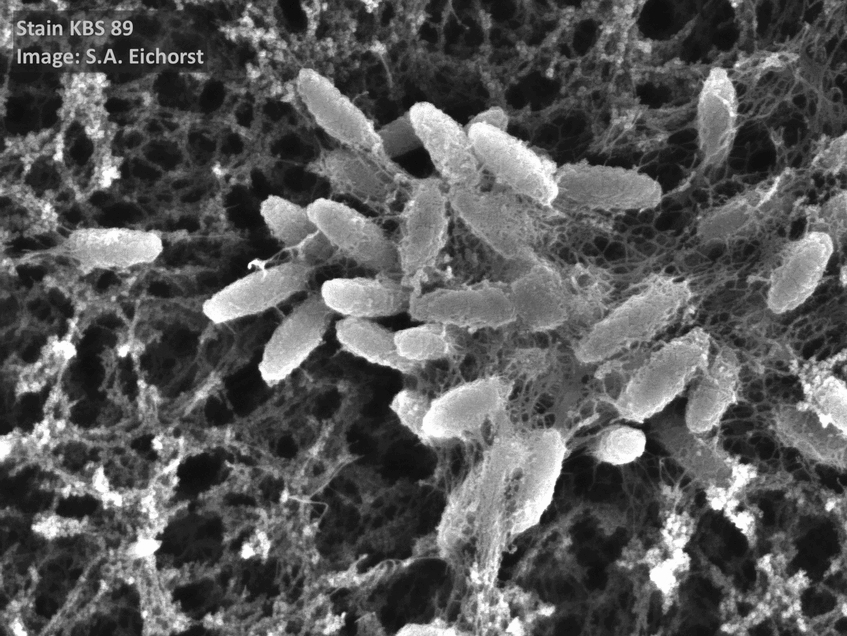
Cultivating the uncultivables
Soil is a plentiful source of microbial life. Yet only a small fraction of the total microbial community has been cultivated. Since my PhD, I have been fascinated with cultivating “yet-to-be-cultivated” microorganisms and find the outcomes of such experiments extremely rewarding as one can better investigate the potential of the microorganism in the laboratory. I have developed novel methods to isolate, detect and subsequently culture elusive soil microorganisms – such as members of the phylum Acidobacteriota, being one of the first researchers to do so.
Single-cell method development for exploring the activity of soil microorganisms
Single-cell method development for exploring the activity of soil microorganisms
One aspect of my research is to investigate the active participants in microbial processes at the single-cell level to confirm their activity and to gain additional information at this scale. As such, we are developing pipelines and are optimizing tools that permit the analysis of our targeted processes down to the single-cell level, such as FISH, NanoSIMS and Raman microspectroscopy combined with stable isotope tracers (such as 13C, 15N and D) in complex systems, such as soil.
Some of our developments have been highlighted by the Joint Genome Institute, Science Highlights: A Single-Cell Pipeline for Soil Samples.

