Physiology And Environmental Importance Of Complete Ammonia Oxidizers (Comammox)

Nitrification, the microbially driven two-step oxidation of ammonia via nitrite to nitrate, is a key process of the biogeochemical nitrogen cycle. Its…
☎ +43 1 4277 91204
✉ holger.daims(at)univie.ac.at
» CV
» u:cris
» u:find

Our research centers on microorganisms that drive the biogeochemical nitrogen cycle, with a particular focus on key nitrifiers: nitrite-oxidizing bacteria (NOB) and complete ammonia oxidizers (comammox). Using a combination of molecular, omics, and microbial physiology approaches, we identify new nitrifying microbes and investigate their metabolism, ecophysiology, and evolution. Our goal is to achieve a fundamental understanding of nitrification in natural and engineered ecosystems, with implications for improving agricultural and wastewater management practices and reducing nitrous oxide emissions.
We are also interested in microbe-microbe interactions within complex microbiomes, such as biofilms in natural and engineered environments. By integrating omics, fluorescence and chemical imaging, and computational analyses, we identify microbial partners and study their interactions in situ. These efforts aim to uncover core principles of microbial interaction that shape the composition, spatial organization, and functional adaptability of microbial communities.
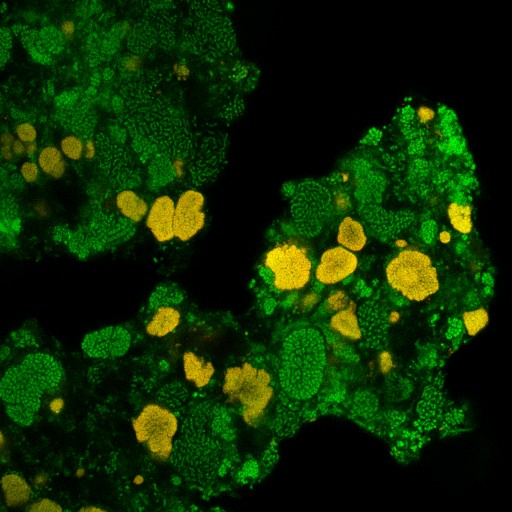
We contributed to the discovery of complete ammonia oxidizers (comammox), microorganisms belonging to the diverse and environmentally widespread genus Nitrospira. Unlike all other known nitrifiers, comammox perform complete nitrification (the oxidation of ammonia to nitrate) within a single organism. This discovery resolved a century-old question in nitrogen cycling and transformed our understanding of this fundamental biogeochemical process. It also opened new research directions on the ecology, evolution, kinetics, regulation, and biochemistry of complete nitrification. Comammox Nitrospira are particularly well adapted to oligotrophic environments: their kinetic traits allow them to outcompete other ammonia oxidizers, including many ammonia-oxidizing archaea, under strong substrate limitation. We investigate the biology of these remarkable organisms using our primary comammox isolate (Nitrospira inopinata), new enrichment cultures, and cultivation-independent approaches in close collaboration with the group of Michael Wagner.
Nitrite-oxidizing bacteria (NOB), which catalyze the second step of nitrification, remain among the most elusive and least understood players in the nitrogen cycle. Our research aims to uncover the biology and ecological roles of NOB in natural habitats and engineered systems such as wastewater treatment plants. We combine (meta)genomic analyses with laboratory experiments to test genome-based hypotheses.
Traditionally viewed as highly specialized with narrow metabolic capacities, NOB are now recognized as remarkably versatile. Our work has revealed that many NOB can oxidize not only nitrite but also alternative substrates such as hydrogen or formate, and can use oxygen or nitrate as terminal electron acceptors. Our research explores how this impressive metabolic flexibility underpins their niche adaptations and survival strategies in diverse and fluctuating environments.
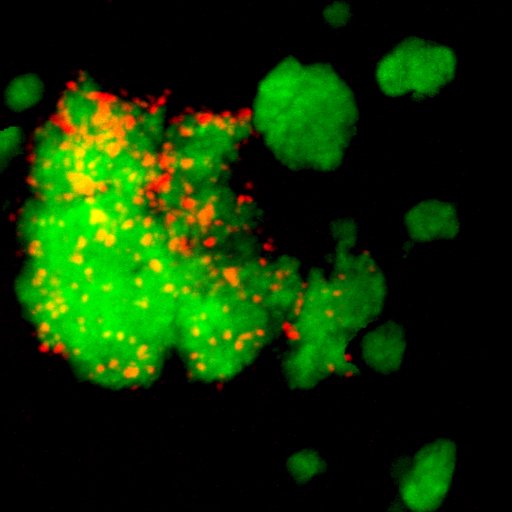
Microbiomes consist of diverse microorganisms that interact in many functionally distinct ways, including the exchange of metabolites, syntrophic substrate degradation, molecular signaling, stress mitigation, competition, and predation. These interactions shape community structure and influence key processes that sustain ecosystem functioning. Our research seeks to unravel how such microbial relationships affect growth, activity, and resilience within microbiomes.
A focus lies on interactions involving nitrifying organisms. As chemolithoautotrophic primary producers, nitrifiers provide organic matter that supports heterotrophic partners, while in turn benefiting from other microbes that supply essential cofactors or alleviate environmental stress. By studying these associations, we aim to understand the ecology of nitrifiers beyond their role in nitrification and to reveal the mechanisms that govern their coexistence and population dynamics within complex microbial communities.
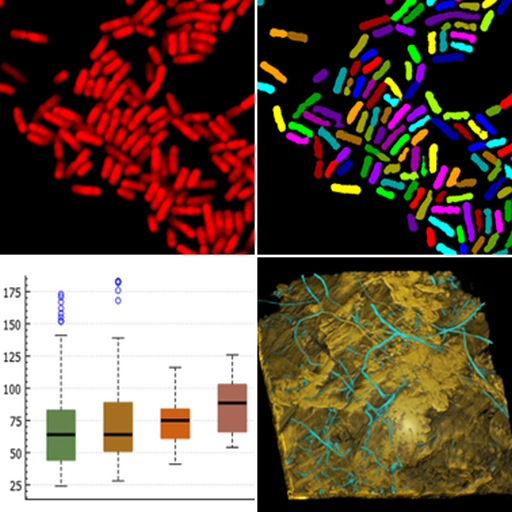
Fluorescence labeling techniques are essential tools in microbial ecology and related microbiological disciplines. A prominent example is fluorescence in situ hybridization (FISH) with rRNA-targeted probes, which enables the cultivation-independent identification, detection, and quantification of microorganisms. Combined with advanced imaging systems such as confocal laser scanning microscopes, FISH allows three-dimensional analyses of labeled microbial populations in biofilms and other spatially complex samples.
The full potential of fluorescence labeling is realized through quantitative digital image analysis. We develop algorithms for the analysis of fluorescence-labeled microorganisms, implemented in our software daime (digital image analysis in microbial ecology). It provides accurate quantification of microbial population and community features in diverse sample types, including biofilms, flocs, and animal or plant tissues. It also offers advanced tools for interactive three-dimensional visualization of confocal image stacks, seamlessly integrated with its quantitative image analysis capabilities.

Nitrification, the microbially driven two-step oxidation of ammonia via nitrite to nitrate, is a key process of the biogeochemical nitrogen cycle. Its…

Nitrification is a key process of the biogeochemical nitrogen cycle and wastewater treatment. Our current knowledge on the involved organisms –…
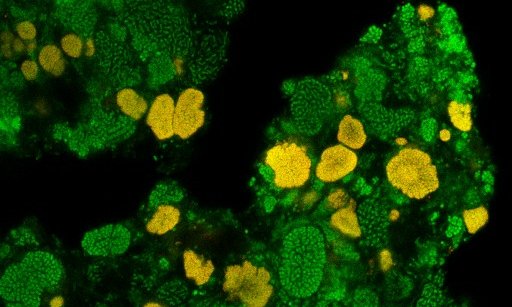
Nitrite-oxidizing bacteria (NOB) are key players of the biogeochemical nitrogen cycle. By catalyzing the second step of nitrification, the oxidation…
Bale NJ, Fujimura H, Pjevac P, Koenen M, Ikeda H, Itagaki S et al. Unusual Plastoquinones in Non-Phototrophic Nitrifying Bacteria. Environmental Microbiology Reports. 2025 Aug 5;17(4):e70174. doi: 10.1111/1758-2229.70174
Kop LFM, Koch H, Speth D, Lüke C, Spieck E, Jetten MSM et al. Comparative genome analysis reveals broad phylogenetic and functional diversity within the order Nitrospirales. ISME Journal. 2025 Jul 22;19(1):wraf151. doi: 10.1093/ismejo/wraf151
Liu T, Duan H, Lücker S, Zheng M, Daims H, Yoan Z et al. Sustainable wastewater management through nitrogen-cycling microorganisms. Nature Water (Nat Water). 2024 Oct 14;2(10):936–952. 1836. doi: 10.1038/s44221-024-00307-5
Palatinszky M, Herbold CW, Sedlacek CJ, Pühringer D, Kitzinger K, Giguere AT et al. Growth of complete ammonia oxidizers on guanidine. Nature. 2024 Sept 19;633(8030):646-653. Epub 2024 Aug 14. doi: 10.1038/s41586-024-07832-z
Bayer B, Liu S, Louie K, Northen TR, Wagner M, Daims H et al. Metabolite release by nitrifiers facilitates metabolic interactions in the ocean. The ISME Journal. 2024 Sept 8;18(1):wrae172. doi: 10.1093/ismejo/wrae172
Holger Daims is part of the FWF-funded Cluster of Excellence (CoE)

