Microscopy and image analysis in microbiology
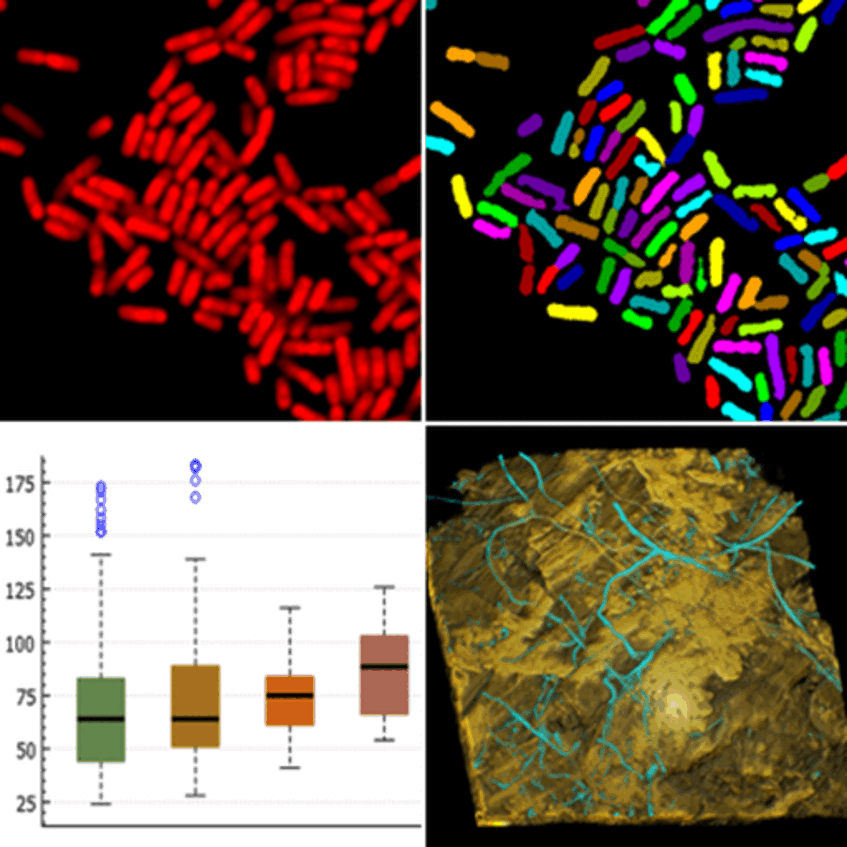
Fluorescence labeling methods are indispensable tools in microbial ecology and other microbiological fields. A prime example is fluorescence in situ hybridization (FISH) with rRNA-targeted probes, which allows for the cultivation-independent identification, detection, and quantification of microorganisms. Advanced instruments like confocal laser scanning microscopes facilitate three-dimensional analysis of fluorescence-labeled target populations in biofilms and other spatially complex samples.
The full potential of fluorescence labeling is realized when quantitative data are derived from micrographs through digital image analysis. We develop algorithms for analyzing fluorescence-labeled microorganisms, incorporated into our software daime (digital image analysis in microbial ecology). This software enables precise quantification of key features in microbial populations and communities across various sample types, including biofilms, flocs, and tissues from animals and plants. It also features tools for interactive 3D visualization of confocal image stacks, fully integrated with its image analysis capabilities.
Website of the daime software
Selected publications on this topic:
- Riva A, Kuzyk O, Forsberg E, Siuzdak G, Pfann C, Herbold C, Daims H, Loy A, Warth B, Berry D. 2019. A fiber-deprived diet disturbs the fine-scale spatial architecture of the murine colon microbiome. Nat. Commun. 10: 4366 https://doi.org/10.1038/s41467-019-12413-0
- Almstrand R, Daims H, Persson F, Sörensson F, Hermansson M. 2013. New methods for analysis of spatial distribution and coaggregation of microbial populations in complex biofilms. Appl. Environ. Microbiol. 79(19):5978-5987 https://doi.org/10.1128/aem.01727-13
- Daims H, Wagner M. 2011. In situ techniques and digital image analysis methods for quantifying spatial localization patterns of nitrifiers and other microorganisms in biofilm and flocs. Methods Enzymol. 496: 185-215 https://doi.org/10.1016/b978-0-12-386489-5.00008-7
- Daims H, Wagner M. 2007. Quantification of uncultured microorganisms by fluorescence microscopy and digital image analysis. Appl. Microbiol. Biotechnol. 75(2):237-248 https://doi.org/10.1007/s00253-007-0886-z
- Daims H, Lücker S, Wagner M. 2006. daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8: 200-213 https://doi.org/10.1111/j.1462-2920.2005.00880.x
