Evolution & environmental diversity of an ancient enzyme and discovery of novel sulfur microorganisms
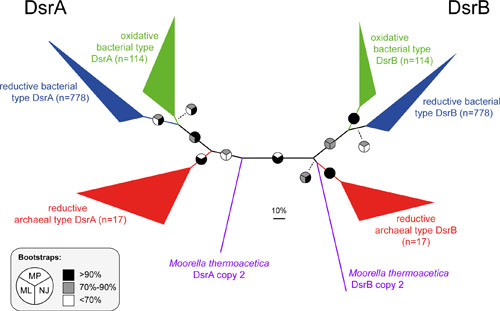
The evolution of DsrAB-type dissimilatory sulfite reductase has been influenced by lateral gene transfer but surprisingly few gene duplication events. This enzyme and its genes have been successfully applied, within certain limits, as diagnostic markers for environmental studies of sulfite/sulfate-reducing and some sulfur-oxidizing microorganisms (Sabehi et al. 2005. PLoS Biol, Loy et al. 2009. Environ Microbiol, Gruber-Vodicka et al. 2011. PNAS). Amplicon sequencing and genome-centric metagenomics of diverse environments has shown that DsrAB-based dissimilatory sulfur metabolism is more widespread among extant bacterial and archaeal phyla than previously recognized (Müller et al. 2015. ISME J, Anantharaman et al. 2018. ISME J). Our publically available dsrAB/DsrAB database, a new multi-level classification system, and new primers also allow for large-scale dsrAB ecology studies using multiplex amplicon sequencing (Pelikan et al. 2016. Environ Microbiol).
Notably, abundant ocean viruses also carry microbial sulfur genes and potentially manipulate the sulfur metabolism of their microbial hosts (Roux et al. 2016. Nature).
Selected publications on this theme:
- Anantharaman K, Hausmann B, Jungbluth SP, Kantor RS, Lavy A, Warren LA, Rappé MS, Pester M, Loy A, Thomas BC, and Banfield JF [2018] Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. doi:10.1038/s41396-018-0078-0
- Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, Poulos BT, Solonenko N, Lara E, Poulain J, Pesant S, Kandels-Lewis S, Dimier C, Picheral M, Searson S, Cruaud C, Alberti A, Duarte CM, Gasol JM, Vaque D, Tara Oceans Coordinators, Bork P, Acinas SG, Wincker P, and Sullivan MB. 2016. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 537: 689–693. doi: 10.1038/nature19366
- Pelikan C, Herbold CW, Hausmann B, Müller AL, Pester M, Loy A. 2016. Diversity analysis of sulfite- and sulfate-reducing microorganisms by multiplex dsrA and dsrB amplicon sequencing using new primers and mock community-optimized bioinformatics. Environ Microbiol. doi: 10.1111/1462-2920.13139
- Müller AL, Kjeldsen KU, Rattei T, Pester M, Loy A. 2015. Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J. 9: 1152-1165.
- Gruber-Vodicka HR, Dirks U, Leisch N, Baranyi C, Stoecker K, Bulgheresi S, Heindl NR, Horn M, Lott C, Loy A, Wagner M, Ott J. 2011. Paracatenula, an ancient symbiosis between thiotrophic Alphaproteobacteria and catenulid flatworms. Proc. Natl. Acad. Sci. USA 108: 12078-12083.
- Loy A, Duller S, Baranyi C, Mußmann M, Ott J, Sharon I, Béjà O, Le Paslier D, Dahl C, Wagner M. 2009. Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur-oxidizing prokaryotes. Environ. Microbiol. 11: 289-299.
- Sabehi G#, Loy A#, Jung KH, Partha R, Spudich JL, Isaacson T, Hirschberg J, Wagner M, Beja O. 2005. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 3: e273. #equal contribution
