Intestinal microbiota - Inflammation and a new method for analysis of microbial physiology in vivo
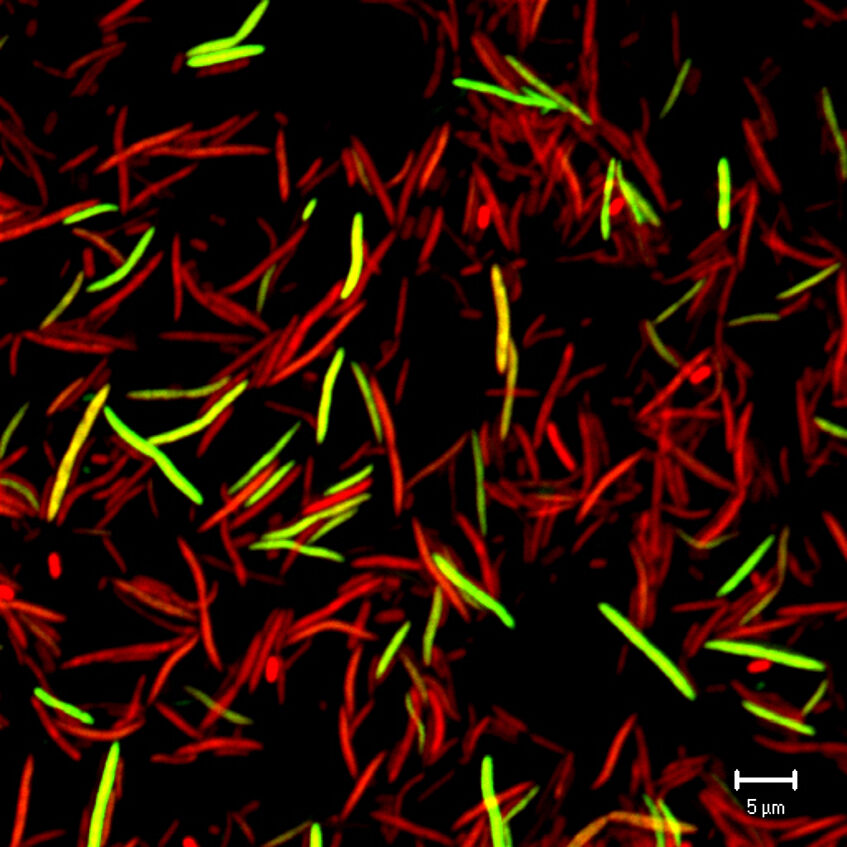
Inflammatory bowel diseases are characterized by an altered gut microbiota composition. By using mouse colitis models, we identified indicator phylotypes for health state (Berry et al. 2012. ISME J, Berry et al. 2015. Front Microbiol) and revealed characteristic changes in the microbial transcriptome and interactions with the host (Schwab et al. 2014. ISME J). In a clinical pilot study, we showed that treatment success and the temporal fecal microbiota dynamics varied among ulcerative colitis patients after receiving fecal microbiota transplantation from healthy donors as an intervention therapy (Angelberger et al. 2013. Am J Gastroenterol). By developing a novel single-cell, isotope-probing method, we could identify, for the first time, important foragers of mouse host-derived protein compounds in vivo and show that the host-foraging phenotype of these bacteria is quantitatively dependent on the complexity of the intestinal microbiota (Berry et al. 2013. PNAS, Stecher et al. 2013. FEMS Microbiol Rev).
Selected publications on this theme:
- Berry D, Kuzyk O, Rauch I, Heider S, Schwab C, Hainzl E, Decker T, Müller M, Strobl B, Schleper C, Urich T, Wagner M, Kenner L, Loy A. 2015. Intestinal microbiota signatures associated with inflammation history in mice experiencing recurring colitis. Front Microbiol. 6: 1408.
- Schwab C, Berry D, Rauch I, Rennisch I, Ramesmayer J, Hainzl E, Heider S, Decker T, Kenner L, Müller M, Strobl B, Wagner M, Schleper C, Loy A, Urich T. 2014. Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J. 8: 1101-1114.
- Berry D, Stecher B, Schintlmeister A, Reichert J, Brugiroux S, Wildd B, Wanek W, Richter A, Rauch I, Decker T, Loy A, Wagner M. 2013. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc. Natl. Acad. Sci. USA 110: 4720-4725.
- Stecher B, Berry D, Loy A. 2013. Colonization resistance and microbial ecophysiology: Using gnotobiotic mouse models and single-cell technology to explore the intestinal jungle. FEMS Microbiol. Rev. 37: 793-829.
- Angelberger S, Reinisch W, Makristathis A, Lichtenberger C, Dejaco C, Papay P, Novacek G, Trauner M, Loy A, Berry D. 2013. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am. J. Gastroenterol. 108: 1620-1630.
- Berry D, Schwab C, Milinovich G, Reichert J, Ben Mahfoudh K, Decker T, Engel M, Hai B, Hainzl E, Heider S, Kenner L, Müller M, Rauch I, Strobl B, Wagner M, Schleper C, Urich T, Loy A. 2012. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 6: 2091-106.
