Univ.-Prof. Dr. Alexander Loy

Alexander Loy’s research covers a diverse spectrum of topics in microbial ecology and symbiosis, from the role of intestinal microbiota in health and disease, to the ecology and evolution of sulfur-compounds-utilising microorganisms. He has long-standing expertise in developing and applying stable isotope-labelling and molecular biology methods for in situ analysis of complex host-associated and environmental microbiota.
Alexander’s group explores how physiological interactions among microbiome members impacts their environment or host. This includes how environmental microbes impact sulfur and carbon cycling, as well as how symbionts and pathogens affect the health and nutrition of their hosts. This knowledge could ultimately translate to development of microbiome-based health interventions. Their current projects focus on microbial sulfur metabolism in wetlands and the intestinal tract, impact of emerging pollutants on human and environmental microbiomes, as well as the development of defined microbial communities for production of new secondary metabolites, such as antibiotics.
Research Topics
Sulfur Microorganisms
Sulfur Microorganisms
It is mainly through microbial activities that the sulfur cycle is intimately intertwined with the cycling of other elements such as carbon or nitrogen. My group is interested in the evolution of sulfur microorganisms, their yet largely unexplored diversity, distribution, and ecological function in various environments, and their roles as symbionts of animals and humans.
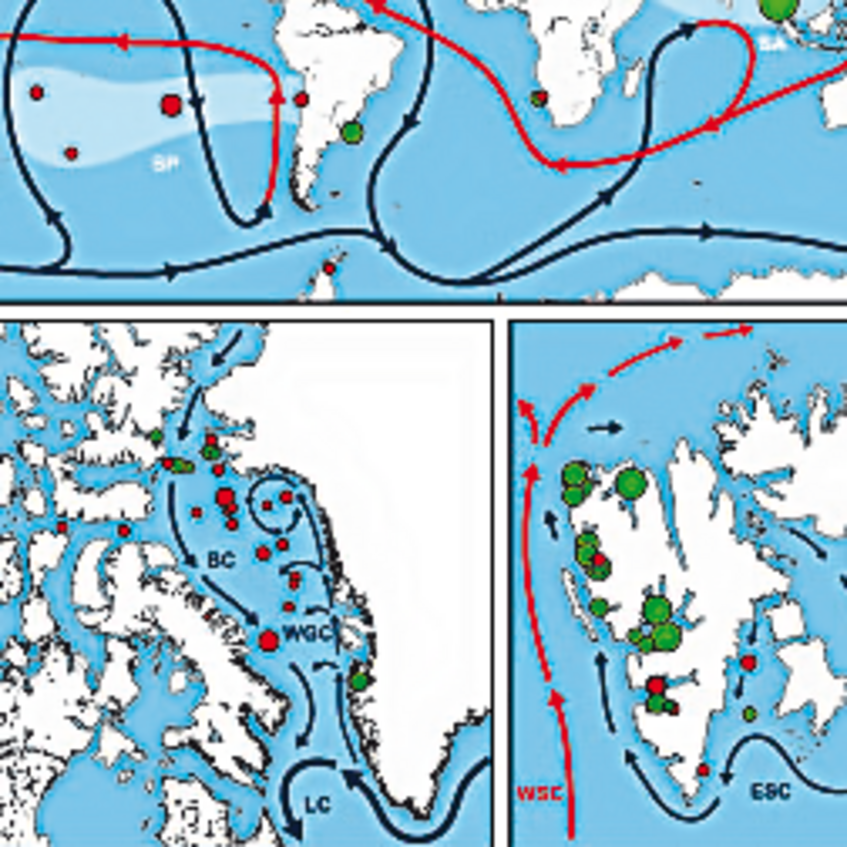
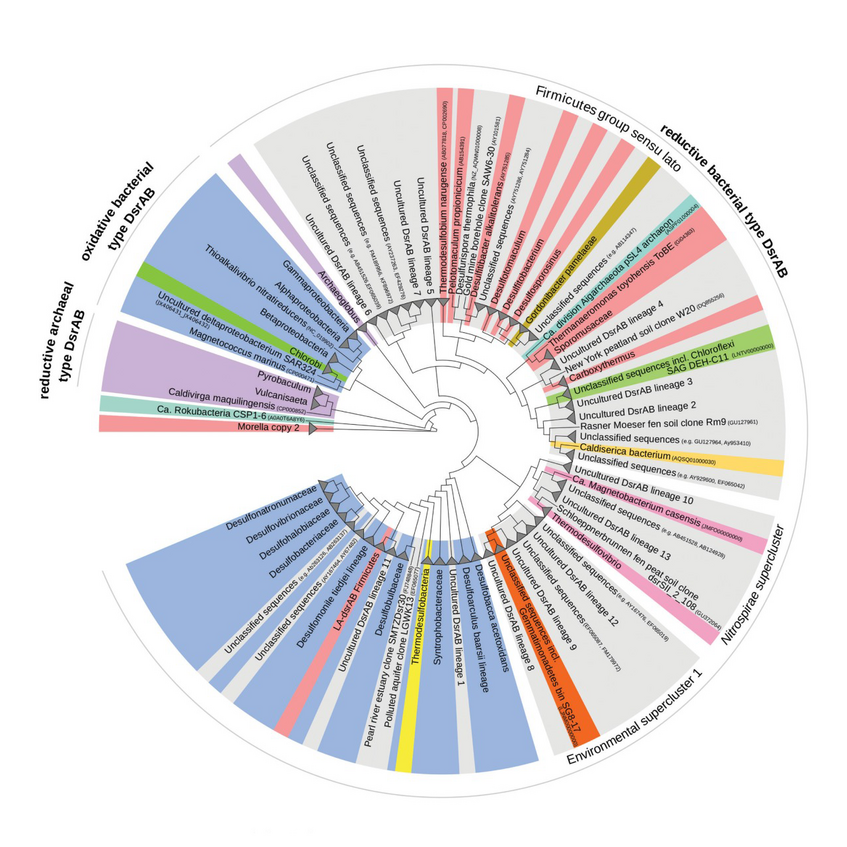
We are investigating the evolution of sulfur microorganisms by comparative phylogenetic analysis of key genes such as dsrAB, encoding dissimilatory (bi)sulfite reductase and by phylogenomics. We specifically aim to reveal the ecology and biogeography of sulfur-cycling microorganisms (e.g. sulfate reducers) in globally relevant anoxic ecosystems such as the ocean floor (Wasmund et al. 2017. Environ Microbiol Rep) and terrestrial wetlands (Pester et al. 2012 Front Microbiol).
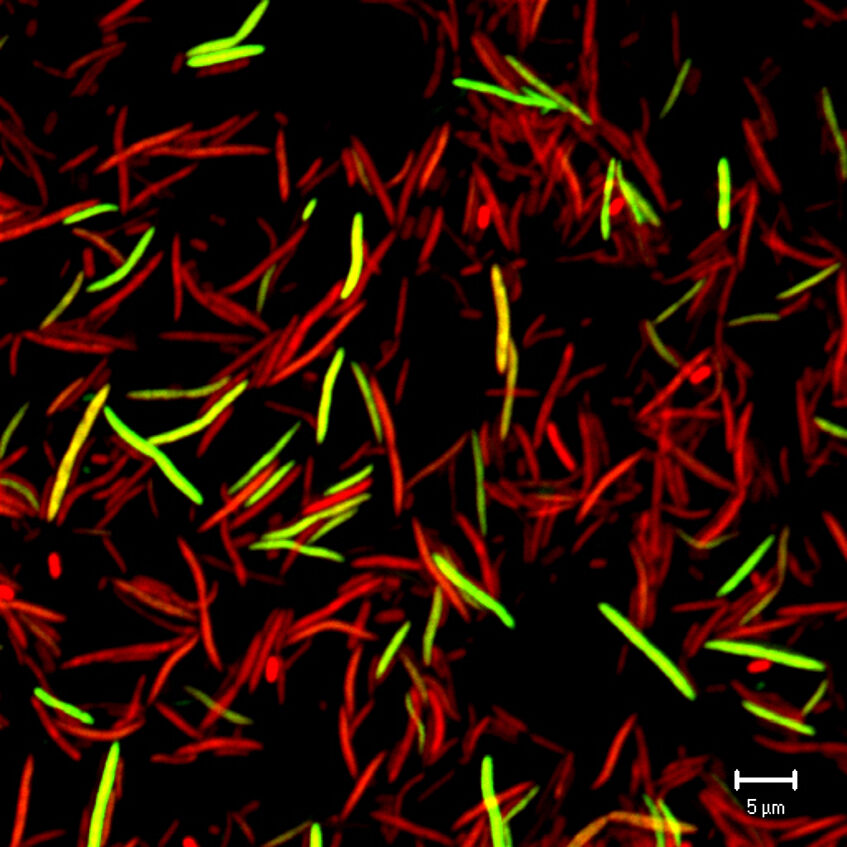
Isotope-labeling techniques allow us to show how the physiological interactions of individual members of complex microbial communities influence important processes in the global carbon and sulfur cycles such as degradation of organic matter and greenhouse gas emissions. We further use genomics and targeted genome-centric gmetagenomic/-transcriptomic approaches to get insights into the genome evolution and metabolism of ecologically important sulfur microorganisms such as rare but cosmopolitan sulfate-reducing Desulfosporosinus species in wetlands (Hausmann et al. 2019. mBio).
Intestinal Microbiota
Intestinal Microbiota
The symbiotic intestinal microbiota provides important benefits to its animal and human host such as breakdown of complex nutritional substrates and protection against pathogens. Changes in the intestinal microbiota are, however, associated with many diseases. We are interested in the ecophysiology of the gut microbiota in health and disease. In the context of various diseases such as inflammatory bowel disease, cancer or intestinal infections, we specifically aim to answer how nutrition impacts the gut microbiota and which/how individual community members influence physiology and health of their host.
Individual cells of the intestinal microbiota either compete for substrates due to functional overlap or cooperate in syntrophic substrate degradation, resulting in a dynamic network of synergistic and antagonistic physiological interactions that are reciprocally influenced by the fluctuating physical and chemical environment of the gut. Here, in particular single cell isotope labeling tools promise yet unavailable detailed insights into host- or diet-derived substrate partitioning and niche competition among individual intestinal microbiota members, including incoming pathogens (Loy and Berry, 2018. Trends Microbiol, Stecher, Berry, and Loy. 2013. FEMS Microbiol Rev).
One focus of our work is the evolution and function of sulfur compounds-utilizing and hydrogen sulfide-producing gut microorganisms because a balanced sulfur metabolism is crucial for intestinal homeostasis and host health. Furthermore, some pathogens such as Salmonella influence the intestinal sulfur metabolism to selectively support their outgrowth and fitness, but the role of the indigenous sulfur microbiota during these intestinal infections is unknown.