Prof. Dr. Dr. h. c. Michael Wagner
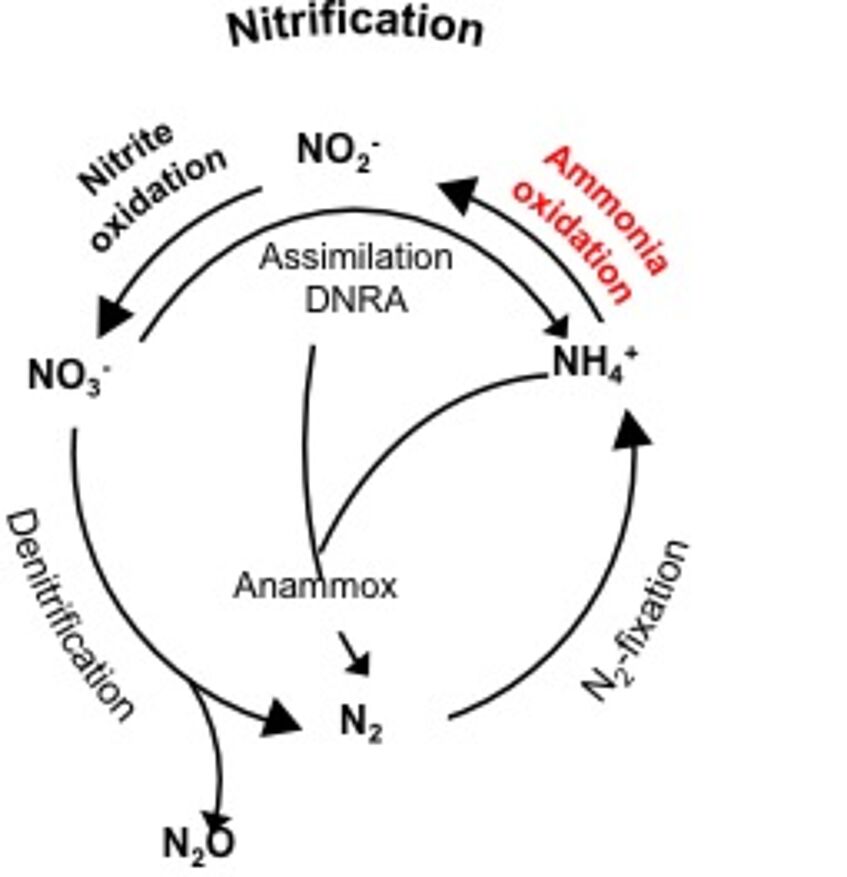
Humans are strongly impacting the global nitrogen cycle by massive use of nitrogen fertilisers. Nitrification leads to fertiliser loss, eutrophication, and greenhouse gas emission, but is essential for efficient wastewater treatment. Research in Michael Wagner‘s group focuses on the ecology, physiology, and evolution of nitrifying microorganisms. Michael Wagner’s group has discovered, cultured, and characterised important new nitrifying bacteria and archaea, including the long sought-after complete nitrifiers, describing unexpected physiological traits in the process.
Michael also has a strong interest in microbial communities driving sewage treatment and in the microbiomes of marine sponges. His group also develop innovative single cell tools to study functional properties of microbes in their natural environment. Michael is an EMBO member, has received an ERC Advanced Grant, the FWF Wittgenstein Award (highest Austrian science award), the Jim Tiedje Award of the International Society for Microbial Ecology, and the Schrödinger Prize of the Austrian Academy of Sciences. He is the director of the FWF Cluster of Excellence “Microbiomes Drive Planetary Heath”.
Ongoing Research Projects
Join the Team
If you are interested in joining our team, explore our open positions and learn more about available PhD and postdoc stipends here.
Teaching
To view Michael Wagner's teaching activities at the University of Vienna, visit u:find.
Group Members
Publications
Strous, M., Pelletier, E., Mangenot, S.
, Rattei, T., Lehner, A., Taylor, M. W.
, Horn, M., Daims, H., Bartol-Mavel, D., Wincker, P., Barbe, V., Fonknechten, N., Vallenet, D., Segurens, B., Schenowitz-Truong, C., Medigue, C.
, Horn, A., Snel, B., Dutilh, B. E., ... Le Paslier, D. L. (2006).
Deciphering the evolution and metabolism of an anammox bacterium from a community genome.
Nature,
440(7085), 790-794.
https://doi.org/10.1038/nature04647
Daims, H., Maixner, F., Lücker, S., Hace, K.
, & Wagner, M. (2006).
Ecophysiology and niche differentiation of Nitrospira-like bacteria, the key nitrite oxidizers in wastewater treatment plants. In
Water Science and Technology: Volume 54 (pp. 21-27). Unknown publisher.
Wagner, M., Nielsen, P. H.
, Loy, A., Nielsen, J. L.
, & Daims, H. (2006).
Linking microbial community structure with function: Fluorescence in situ hybridization-microautoradiography and isotope arrays.
Current Opinion in Biotechnology,
17(1), 83-91.
http://www.scopus.com/scopus/inward/record.url?eid=2-s2.0-31944434615&partner@&rel=R5.0.4