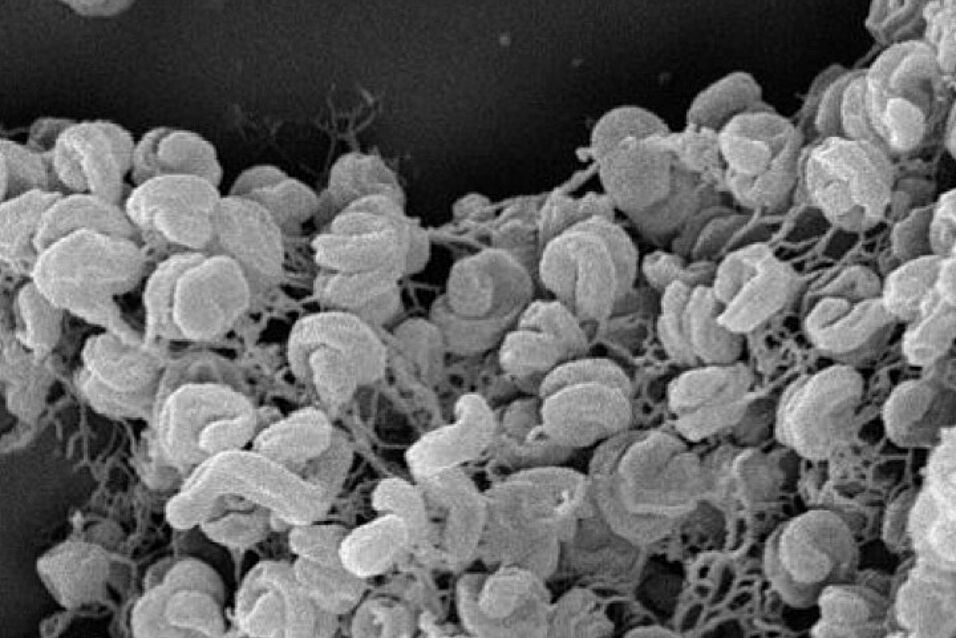
This project will use a full-scale wastewater treatment plant as model ecosystem to study the microdiversity of uncultured NOB, which coexist in the same activated sludge, in a concerted approach comprising in-depth comparative genomics, population genetics, and in situ single-cell physiology. It builds upon a novel method to identify, physically extract, and genome-sequence individual cell aggregates (microcolonies) of nitrifiers from activated sludge by Raman microspectroscopy, laser tweezing, and single-cell genomics. Population genetic analyses will be performed on the obtained NOB genomes to resolve the fine-scale population structure of microdiverse NOB.
Genome-based hypotheses on (possibly novel) niche-defining metabolic features of NOB clades will be tested in situ with the same living NOB populations. We will apply a battery of single-cell tools such as FISH with a high phylogenetic resolution, isotope labeling, Raman microspectroscopy, microautoradiography, and NanoSIMS analyses. Symbiotic AOB-NOB interactions will be characterized by genomics, in situ experiments, and spatial co-aggregation analyses. The genomes of microbial predators or other heterotrophic symbionts, which were attached to the extracted microcolonies, will be co-sequenced with the NOB genomes and will inform on possible interactions of microdiverse NOB with non-nitrifiers.
This project is funded by the FWF - Austrian Science Fund (P27319-B21).
Investigated by: