What are they doing there? New single cell tools for functional analyses of microbes in their ecosystems
My team has a long-standing interest in the development of methods for functional analyses of microbes within complex microbial communities (Wagner et al. 1998; Adamczyk et al. 2003). For example, we pioneered the combination of FISH and microautoradiography (Lee et al., 1999) that enabled microbial ecologists for the first time to observe substrate utilization of uncultured individual microbial cells in their natural habitat. Currently, second generation methods for single cell isotope probing mainly using 13C-, 15N and 2H-labeled compounds are developed and combined with single cell genomics approaches. For the detection of isotopes within microbial cells we use nanometer-scale secondary ion mass spectrometry (NanoSIMS) and Raman microspectroscopy.
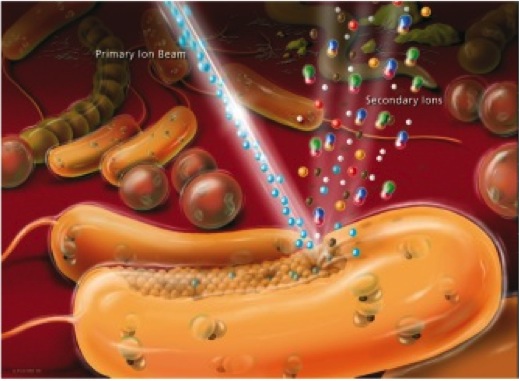
NanoSIMS. NanoSIMS imaging is perfectly suited to measure and visualize the distribution of virtually any elements and their stable isotopes of interest in microbial cells. We run in our team since 2010 a CAMECA NanoSIMS 50L (the only NanoSIMS instrument in Austria) that offers a spatial resolution for element/ isotope mapping down to 50 nm and thus even allows highly sensitive analyses at the sub-cellular level. The NanoSIMS 50L is equipped with Cs+ and O- primary ion sources, an electron gun for analysis of insulating samples, a secondary electron detector, and a magnetic sector mass analyser with a large version of the magnet and a multi-collection system of 7 detectors all equipped with Faraday cups and electron multiplier detectors. In microbial ecology we combine NanoSIMS with stable isotope probing and cell identification techniques such as fluorescence in situ hybridization to obtain previously inaccessible information about the functional role of microorganisms in their environment. Using this approach, previously unrecognized physiological properties of bacteria and archaea thriving in soils, microbial mats, deep groundwater samples and within corals as well as mice guts could be deciphered.
Now working on this theme: Arno Schintlmeister
Selected publications on this theme:
Berry D, Stecher B, Schintlmeister A, Reichert J, Brugiroux S, Wildd B, Wanek W, Richter A, Rauch I, Decker T, Loy A, Wagner M. 2013. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc. Natl. Acad. Sci. USA 110: 4720-4725.
Koch H, Galushko A, Albertsen M, Schintlmeister A, Gruber-Dorninger C, Lücker S, Pelletier E, Le Paslier D, Spieck E, Richter A, Nielsen PH, Wagner M, Daims H. 2014. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science 345: 1052-1054.
Woebken D, Burow L, Behnam F, Mayali X, Schintlmeister A, Fleming E, Prufert-Bebout L, Singer S, López Cortés A, Hoehler T, Pett-Ridge J, Spormann A, Wagner M, Weber P, Bebout B. 2015. Revisiting N2 fixation in Guerrero Negro intertidal microbial mats with a functional single-cell approach. ISME J. 9: 485-496.
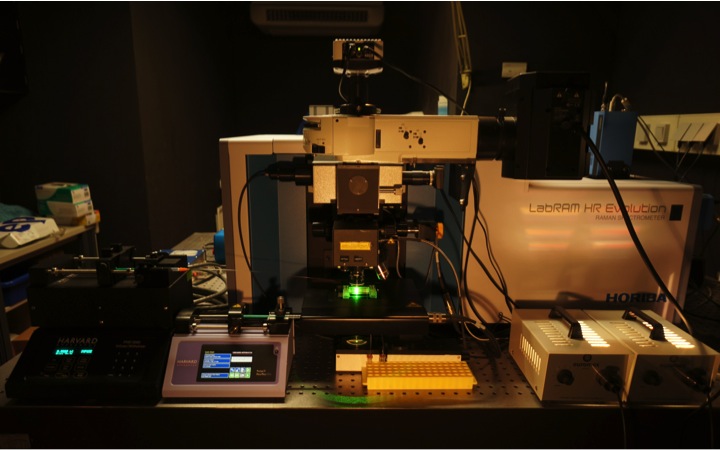
Raman microspectroscopy. In my lab we develop new confocal Raman microspectrocopy-based methods for functional analyses of microbes in complex ecosystems. Raman microspectroscopy has single cell resolution and is nondestructive. It enables us to record within seconds a chemical fingerprint of a microbial cell that reveals the presence of defined storage compounds (Milucka et al., 2012), cytochromes and pigments. Raman microspectroscopy can be directly combined with FISH (Huang et al. 2007) for simultaneous identification of the analyzed microbes. Furthermore, Raman microspectroscopy can be applied to detect and quantify the incorporation of stable isotopes in individual microbial cells and is the most straightforward technique to perform single cell stable isotope probing experiments with complex microbial communities. In addition to detection of 13C-labeled cells (Huang et al. 2007), we recently applied Raman microspectroscopy for tracking the incorporation of deuterium from heavy water in microbial cells in order to measure their activity (Berry et al. 2015). Excitingly, Raman spectra of microbial cells can also be recorded while holding them with an optical tweezer. Subsequently, cells can then be sorted according to their Raman spectrum for single cell genomics or cultivation (Berry et al. 2015). In collaboration with Roman Stocker (ETH, Switzerland) we have developed a microfluidics chamber for high-throughput sorting of microbial cells according to their Raman spectra for directly combining single cell stable isotope probing and single cell genomics or cultivation (Lee et al. 2019).
At DOME two cutting edge confocal Raman microspectrometer are available. A LabRAM HR800 and an HR Evolution (both from Horiba Jobin-Yvon). Availabe lasers are a 532-nm neodymium-yttrium aluminium garnet laser, a pulsed 532-nm laser, a 785 nm laser, and a 1,064-nm laser for optical trapping.
Now working on this theme: Márton Palatinszky, Markus Schmid, (previously Tae Kwon Lee, Christoph Böhm, Esther Mader)
Selected publications on this theme:
Berry D, Mader E, Lee TK, Woebken D, Wang Y, Zhu D, Palatinszky M, Schintlmeister A, Schmid MC, Hanson BT, Shterzer , Mizrahi I, Rauch I, Decker T, Bocklitz T, Popp J, Gibson CM, Fowler PW, Huang WE, Wagner M. 2015. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl. Acad. Sci. USA Jan 13;112(2):E194-203.
Huang WE, Stoecker K, Griffiths R, Newbold L, Daims H, Whiteley AS, Wagner M. 2007. Raman-FISH: Combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ. Microbiol. 9: 1878-1889.
Milucka J, Ferdelman TG, Polerecky L, Franzke D, Wegener G, Schmid M, Lieberwirth I, Wagner M, Widdel F, Kuypers MMM. 2012. Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature 491: 541-546.
Lee KS, Palatinszky M, Pereira FC, Nguyen J, Fernandez VI, Mueller AJ, Menolascina F, Daims H, Berry D, Wagner M, Stocker R. 2019. An automated Raman-based platform for the sorting of live cells by functional properties. Nat Microbiol 6: 1035-1048.
