daime: Features
Image import
daime imports single images, image series, and 3D z-stacks from manufacturer formats including Leica LIF, Zeiss LSM, and many others. It also supports image import from standard TIFF files. Accepted formats include 8-, 12-, and 16-bit grayscale and 24-bit RGB color images.
Background reduction
Various methods for background and noise reduction and illumination correction can be applied to improve images for downstream processing.

Pre-processing tools
Filters like edge detection, histogram stretching, morphological and boolean operations are available for image analysis and visualization workflows - all in 2D and 3D.

Segmentation
2D and 3D image segmentation tools include global and local intensity thresholding by various methods, edge detection, object splitting, and color-based object recognition.
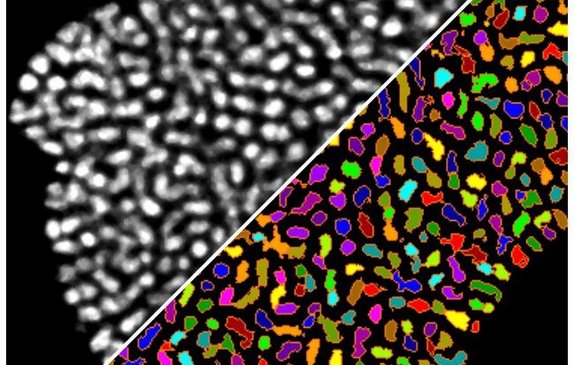
Object features & classification
Measure object features (size, shape, brightness, color, localization, and more) for flexible classification. Each object can belong to multiple classes, enabling advanced and versatile image analyses.

Abundance quantification
Absolute and relative abundances of microbial populations are determined by cell counting or by quantifying biovolume fractions using an efficient stereological method.

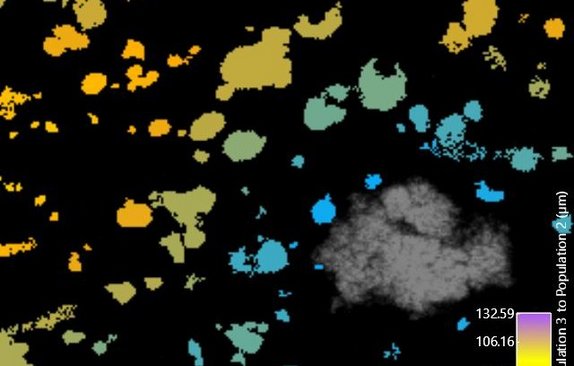
Spatial pattern analysis
Spatial localization patterns of microbial biofilm populations are quantified in 2D and 3D by methods from stereology and spatial statistics.
Biofilm virtual sectioning
Special tools slice biofilms in 2D images and 3D z-stacks in any direction, generating virtual sections or grids for analyzing biofilm communities in defined spatial regions and depth layers.

Analyses with reference objects
Quantify population features in relation to other objects: for example, count endosymbionts within host cells or measure fluorescence intensity of labeled cells in different regions of a large cell aggregate.

FISH probe evaluation
Dissociation curve analysis of rRNA-targeted FISH probes from a formamide series to determine the optimal hybridization stringency.

Visualization
High-quality interactive 3D visualization of z-stacks: explore from any perspective with fly-through, illumination, shadows, stereo rendering, 3D clipping, keyframe-based animation, movie export, and more - all fully integrated with the other program features.

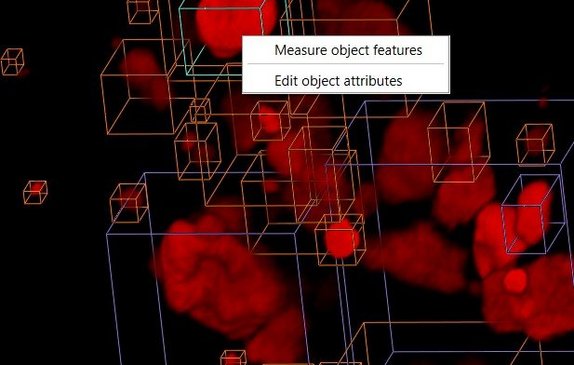
Integrated GUI
A convenient graphical user interface integrates all functions. For example, the Object Editor lets you interactively define, measure, classify, split, merge, colorize, and select 2D or 3D objects for downstream analyses - and much more!
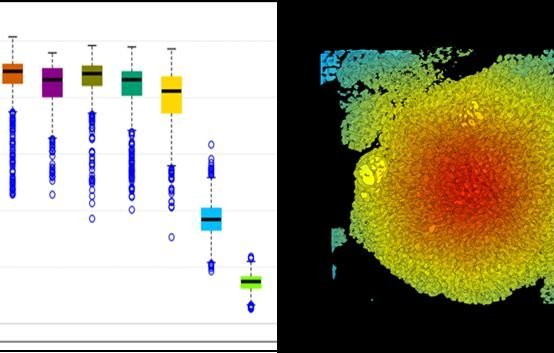
Data presentation
Analysis results appear as data tables and plots that can be browsed on-screen by scrolling, panning, and zooming. Results of many analyses can also be shown as 2D or 3D heatmaps mapped onto image data, fully compatible with all visualization options in daime.
Interoperability
Export images, analysis results, and plots for use in other software, or import object definitions from external segmentation tools. daime also interacts with R: export data in R-ready formats for advanced statistical analyses or auto-generate R scripts for plotting measured data.
Reproducibility
All actions are automatically logged and stored with image data - an integrated lab journal for image analysis! Users can tag images, analyses, and results with comments. Complete image datasets, analysis tasks, results, and visualization sessions can be saved in a project file for easy cross-platform sharing.
...and more!
daime is a feature-rich software designed to help you get the most from your microbial fluorescence image data. Download the program, explore the documentation and tutorial, and give it a try!