Dr. Daan Speth
Senior Scientist at the Centre for Microbiology and Environmental Systems Science

Research Profiles
Social Media
Publications Overview
Recent advances in sequencing technologies have dramatically altered our view of microbial diversity, and led to major discoveries of novel organisms involved in the carbon and nitrogen cycles. During his PhD at Radboud University (Netherlands), Daan applied metagenomics and other sequence-based bioinformatics to study microbial communities in wastewater and marine environments, with a focus on nitrogen and carbon transformations. He continued this work at Caltech (USA) and the Max Plank Institute of Marine Microbiology (Germany), and additionally became fascinated by the evolutionary history and diversity of protein families essential for anaerobic microorganisms. Since 2023, he has continued working on nitrogen cycling organisms as well as protein evolution at the University of Vienna.
Research Topics
Genomics and physiology of nitrifiers
Nitrifying microorgansisms that convert ammonium to nitrite and nitrate play a key role in the global biogeochemical cycle of nitrogen. Nitrifiers present both opportunities and challenges to humans. On one hand, we apply these organisms for our benefit in wastewater treatment, but on the other we want to inhibit their role in fertilizer loss from agricultural fields. Understanding the physiology of nitrifiers and their role in diverse ecosystems is a central goal of the nitrification group. I approach this challenge primarily through the lens of (comparative) genomics, integrating knowledge and generating hypotheses to test in collaboration with colleagues in the group.
Data driven physiology: molybdoenzymes
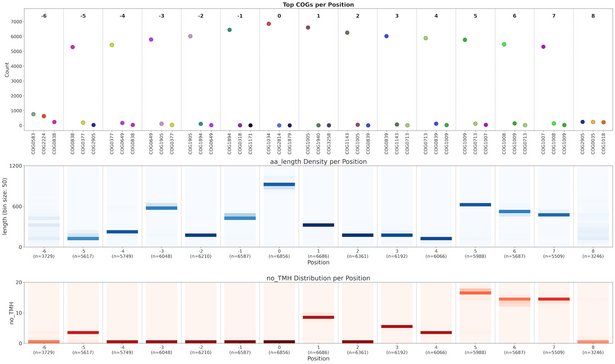
The genomic era has generated millions of genomes of Bacteria and Archaea, belonging to over 300,000 species. For more than 85% of those species, the only information we have is their genome sequence. Interpreting the physiology and ecology of these organisms is a major challenge in microbiology. I tackle this challenge using data-driven physiology. Combining and integrating global scale data sources to prioritize protein complexes for experimental characterization. The current focus of this approach are molybdoenzymes of the Mo/W bis PGD superfamily. These protein complexes catalyze a wide range of biochemical reactions, and are involved in the biogeochemical cycles of carbon, nitrogen, sulfur, iodine, selenium, and arsenic.
GlobDB genome database
Genomes from metagenomes are giving unprecedented insight in the distribution and evolutionary history of Bacteria and Archaea. I built and maintain the GlobDB, the most comprehensive database of species dereplicated microbial genomes currently available. The GlobDB includes a full 7-level taxonomy that extends the established taxonomy of the genome taxonomy database (GTDB) and resources for sequencing dataset classification. Genome annotations and anvi'o databases are available for all 306,260 GlobDB genomes. We have generated protein language model embeddings for a clustered subset of 80 million proteins. All GlobDB resources are available for download at https://globdb.org/

Bioinformatics tool development
To add additional value to the GlobDB, I'm developing the amino acid sequence toolkit (AASTK) together with Marcel Rennig. This toolkit enables the creation of amino acid sequence datasets of GlobDB proteins to be used in further analyses. It integrates previously published approaches for protein alignment score ratio analyses and alignment score matrix clustering, as well as a novel approach for genomic context analysis. AASTK is in active development at https://github.com/dspeth/aastk. With Ben Coltman, I am co-developing MetaCoOc, a tool to detect co-occurence of microbes in metagenome data, which is available at https://github.com/bcoltman/metacooc.
Team Members
- Marcel Rennig | Master's Student + IT Support
- Lisa Bauer | Master's Student
Publications
Data Reuse Consortium, Wagner M, Rattei T, Osvatic J, Speth D. A roadmap for equitable reuse of public microbiome data. Nature Microbiology. 2025 Sept 26;10(10):2384-2395. Epub 2025 Sept 26. doi: 10.1038/s41564-025-02116-2
Kop LFM, Koch H, Speth D, Lüke C, Spieck E, Jetten MSM et al. Comparative genome analysis reveals broad phylogenetic and functional diversity within the order Nitrospirales. ISME Journal. 2025 Jul 22;19(1):wraf151. doi: 10.1093/ismejo/wraf151
Nan Q, Speth D, Qin Y, Chi W, Milucka J, Gu B et al. Biochar application using recycled annual self straw reduces long-term greenhouse gas emissions from paddy fields with economic benefits. Nature Food. 2025 May;6(5):456-465. Epub 2025 Mar 12. doi: 10.1038/s43016-025-01124-z
Glasl B, Kitzinger K, Luter HM, Legin A, Salas Hernandez EK, Heldwein N et al. Branched-chain amino acid assimilation promotes mixotrophy of ammonia-oxidizing archaeal sponge symbionts. bioRxiv. 2025. doi: 10.1101/2025.09.09.672702
Peña-Salinas ME, Speth DR, Utter DR, Spelz RM, Lim S, Zierenberg R et al. Thermotogota diversity and distribution patterns revealed in Auka and JaichMaa 'ja 'ag hydrothermal vent fields in the Pescadero Basin, Gulf of California. PeerJ. 2024 Aug 19;12(8):e17724. doi: 10.7717/peerj.17724